Hubert and Arabie's Adjusted Rand
Adjusted Rand (Hubert & Arabie, 1985) should be used to access the global congruence of two typing methods. It gives the overall concordance of two methods taking into account that the agreement between partitions could arise by chance alone.
where:
N represents the total sample size, ni the number of species belonging to the cluster i of partition A, and nj the number of species belonging to the cluster j of partition B.
It can be specially interesting to determine cut-off values for dendrograms where it maximizes the congruence with the method involving the dendogram information and any other method.
However this coefficient can take low values in the case if a method, being more discriminatory that other, further refines the clusters (groups) formed by another. As an example, if you use the Microbial Typing Test Data you can see that the Adjusted Rand value for the agreement between the groups formed by PFGE Sma80 (groups formed by a cut-off at 80% similarity in a Dice/UPGMA analysis of Pulsed-field Gel Electrophoresis band patterns of the isolates digested with SmaI) and T type is 0.566, which is rather low.
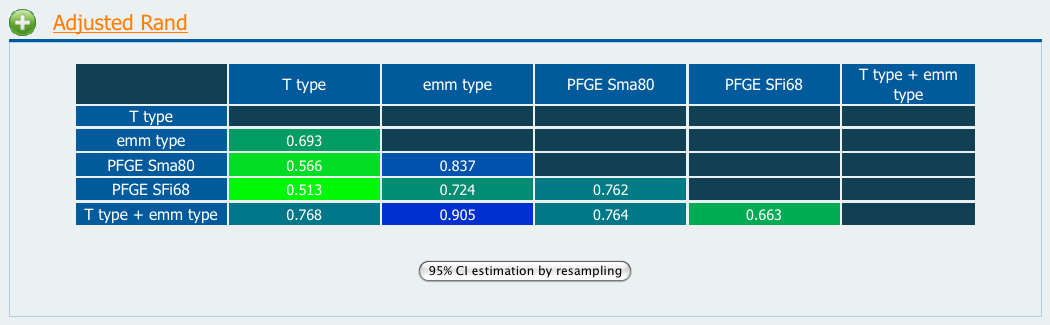 Figure 1
Figure 1The 95% confidence intervals can be calculated by resampling (see Jackknife CI) by clicking the button bellow the results' table.
The confidence interval allows the comparison of different Adjusted Rand values for a single method to see which method is more congruent with the one being tested. As an example, one can determine which methods are globally more congruent with emm type by analyzing the resulting AR values and respective jackknife pseudo-values confidence intervals in the Microbial Typing Data example: T type and PFGE Sfi68 are the less congruent with emm type of the all the methods analyzed (T type 95% CI:0.621<AR<0.767; PFGE Sif68 95% CI:0.653<AR<0.797). Nevertheless we can not exclude the hypothesis, at 95% confidence level, that PFGE Sma80 (PFGE Sma80 95% CI:0.776<AR<0.900) is as congruent with emm typing as PFGE Sfi68 (since their CIs overlap).
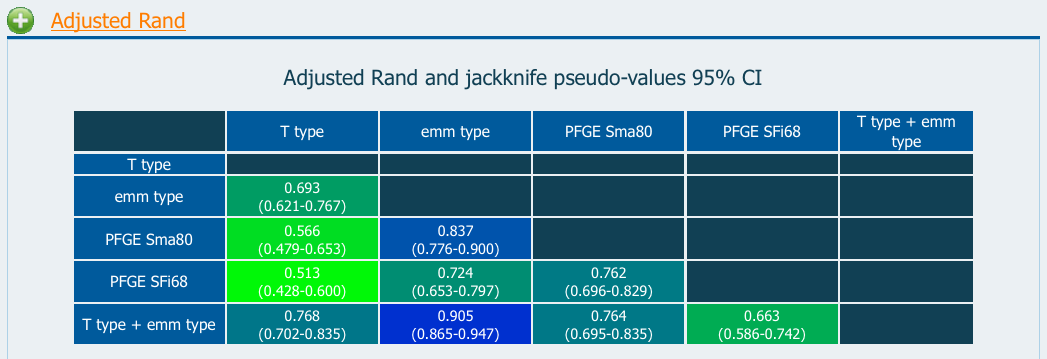 Figure 2
Figure 2
